Introduction:
OBSM is a pipeline calculate Optimal Branch Specific Model with CODEML (belong to PAML package). It is written by Perl. OBSM will calculate a bunch of branch model based on Dynamic Programing prediction. Accoridng to our analysis, OBSM will logically find out the optimal model--in 50 cases we recalculate, OBSM find out much better models in 47 times, while the remain 3 times are the same model compare to the previous study.
Citation:
Zhang, C., et al., Dynamic programming procedure for searching optimal models to estimate substitution rates based on the maximum-likelihood method. Proc Natl Acad Sci U S A, 2011. 108(19): p. 7860-5.
Related Program:
This program require CODEML (belong to PAML package) to get the results. The newst version of PAML please click HERE. Since our pipeline was design earlier (2011), you can
Download
the old version to fit format.
Data Preparation:
- check the codeml in the right path
replace "/home/chengjun/PAML/paml43/bin/codeml" to correct path
six places need to replace in M1_V022.pl
two places need to repalce in M2_V022.pl
two places need to repalce in M3_V022.pl
- set the CPU processor number
my $processor="8"; in M1_V022.pl
- set threshold k in M3_V22.pl, the defalt vlaue is 0.5
- this version is runing well on linux OS
Make sure:
1. before you running the program, make sure the tree file (gene_tree.trees) was one line only (especially in method 2 and 3)
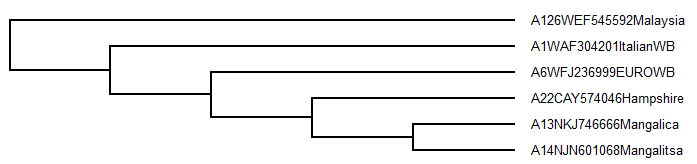
e.g. (((A6WFJ236999EUROWB,(A22CAY574046Hampshire,(A13NKJ746666Mangalica,A14NJN601068Mangalitsa))),A1WAF304201ItalianWB),A126WEF545592Malaysia);
2. when you running the method 1, please make sure the gene_tree.trees and dna_seq_for_paml.txt was in a doucument (we suggest only this two files was in this doucument), and the script file OBSM_Download_M1_V022.pl should be in the same path with the document(not with two files).
--------OBSM_Download_M1_V022.pl
--------method1
++++-------gene_tree.trees
++++-------dna_seq_for_paml.txt
e.g.

3. when you running the method 2 and 3, keep gene_tree.trees, dna_seq_for_paml.txt, codeml.ctl and OBSM_Download_M2_V022.pl (or OBSM_Download_M3_V022.pl) in one document
Get Results:
1. for method 1, please run the script "get_optimal_value_of_methodI.pl" out the document, keep it with "OBSM_Download_M1_V022.pl" at the same path.
+++++++++++++++++++++++++++for example+++++++++++++++++++++++++++++++++++
$ls
get_optimal_value_of_methodI.pl method1 OBSM_Download_M1_V022.pl
$cd method1
$ls
gene_tree.trees dna_seq_for_paml.txt
$cd ..
$perl OBSM_Download_M1_V022.pl
...runing...
$cd method1
$ls
ORM FRM TRM-0 TRM-1 ... 2-RM 3-RM ... gene_tree.trees dna_seq_for_paml.txt last_rel.txt
$cd ..
$perl get_optimal_value_of_methodI.pl
Models
P-value
ORM VS TRM
4.25403991908269e-05
**
3-RM VS TRM
1.2053640994325e-05
**
4-RM VS 3-RM
0.0189298977228625
*
5-RM VS 4-RM
0.772709842396476
6-RM VS 4-RM
0.282626511596699
7-RM VS 4-RM
0.16012875572965
8-RM VS 4-RM
0.158534461451702
9-RM VS 4-RM
0.191932481819565
10-RM VS 4-RM
0.28029278277388
11-RM VS 4-RM
0.264839501321693
12-RM VS 4-RM
0.29512101692376
13-RM VS 4-RM
0.347358105298401
14-RM VS 4-RM
0.387349935595905
15-RM VS 4-RM
0.313549671853572
16-RM VS 4-RM
0.147312906212141
17-RM VS 4-RM
0.183966606307003
18-RM VS 4-RM
0.249076686890239
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
2. for method 2 and 3, please run the script in the same document,
++++++++++++++++++++++++++++++++for example++++++++++++++++++++++++++++++++++++
$ls
method2
$cd method2
$ls
codeml.ctl dna_seq_for_paml.txt OBSM_Download_M2_V022.pl gene_tree.trees get_optimal_value_of_methodII.pl
$perl OBSM_Download_M2_V022.pl
...runing...
$ls
codeml.ctl dna_seq_for_paml.txt OBSM_Download_M2_V022.pl gene_tree.trees all_rel.txt
2_ratio_models 3_ratio_models 4_ratio_models ...
$vi all_rel.txt
$perl get_optimal_value_of_methodII.pl
please input the result file name of method II
-1069.136623
-1064.996311
-1060.927361
-1058.527587
-1057.335090
-1056.200276
-1055.894501
-1055.299100
0.004(1 *)
0(2 *)
0(3 *)
0(4 *)
0(5 *)
0(6 *)
0(7 *)
0.004(1 *)
0.002(2 *)
0.002(3 *)
0.001(4 *)
0.003(5 *)
0.004(6 *)
0.028(1 *)
0.028(2 *)
0.024(3 *)
0.039(4 *)
0.047(5 *)
0.123(1)
0.098(2)
0.153(3)
0.168(4)
0.132(1)
0.237(2)
0.254(3)
0.434(1)
0.406(2)
0.275(1)
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++