News
2022-04-14
R-loopAtlas v2.0 (A big innovation) has been onlined !
Abundant R-loop data and deepRloopPre come online!2018-12-25
R-loopAtlas v1.0 (First Open Version) has been released !
Visitors
Introduction
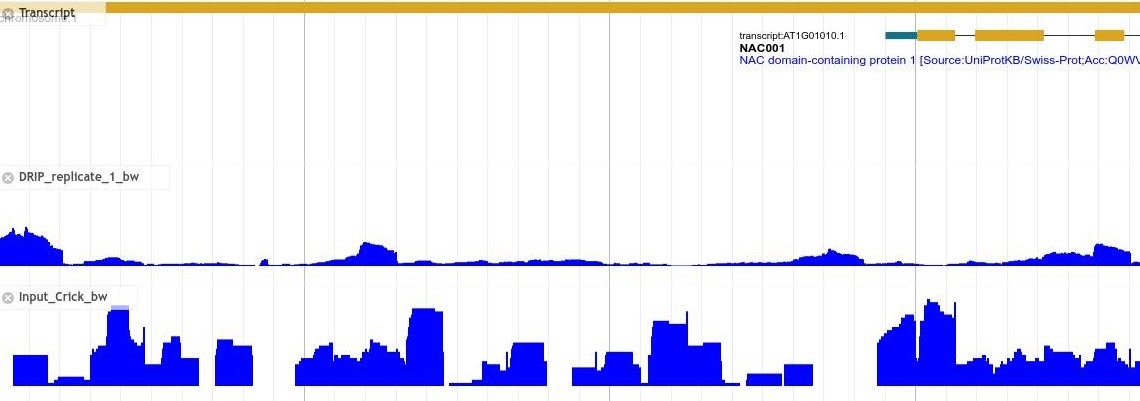
R-loop, three-stranded nucleic acid structures composed of one strand RNA: DNA hybrid and another strand DNA single-stranded, plays an important role in many physiological and pathological processes. Due to its importance, the genomic localization and regulatory functions of R-loop have aroused great interest among researchers. However, there is no similar database and prediction tool to support the R-loop research in plants. To cope with the challenge, we developed deepRloopPre and R-loopAtlas.
R-loopAtlas
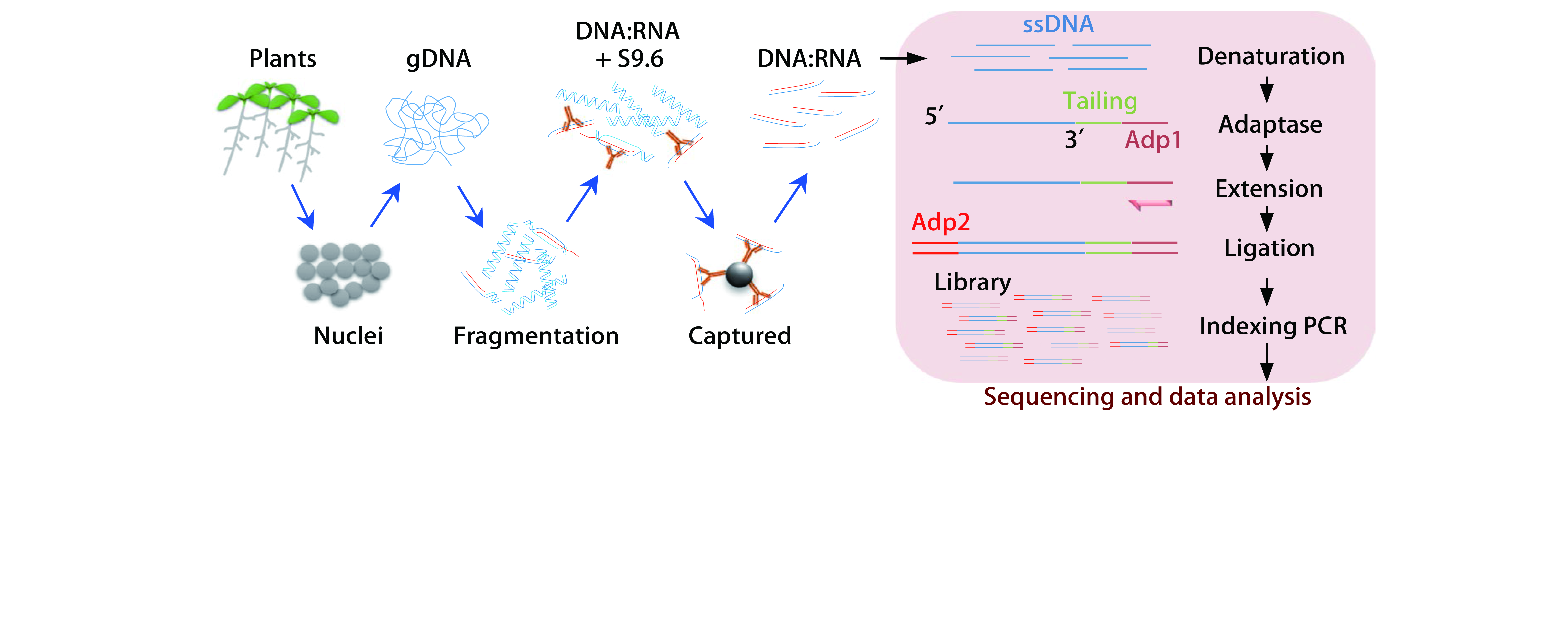
R-loopAtlas recorded R-loop data for 4 species obtained by 3 experimental techniques, and R-loop data for 254 species predicted by deepRloopPre using 4 models, among which the R-loop data of Arabidopsis thaliana is obtained by ssDRIP-seq and Karanyi DRIP-seq, respectively, the R-loop data of Oryza sativa is obtained by ssDRIP-seq and Fang DRIP-seq, respectively, the R-loop data of Zea mays and Glycine max are obtained by ssDRIP-seq, and the R-loop data of 254 plant species are predicted by four deepRloopPre models (the model trained with A. thaliana, with extended A. thaliana, with O. sativa, and with D. rerio, respectively).
deepRloopPre
deepRloopPre is a powerful deep learning tool and could predict the locations and profiles of strand-specific R-loops, with better performance compared to other tools in plants.
If you use R-loopAtlas, please cite: Li, K., Wu, Z., Zhou, J., Xu, W., Li, L., Liu, C., Li, W., Zhang, C., & Sun, Q. (2022). R-loopAtlas: An integrated R-loop resource from 254 plant species sustained by a deep-learning-based tool. Molecular plant,S1674-2052(22)00447-6.