Tutorial
Here, we provide a step-by-step tutorial on how to use our R-loopAtlas. It makes easier access to our database by reading this section.
Overview of R-loopAtlas
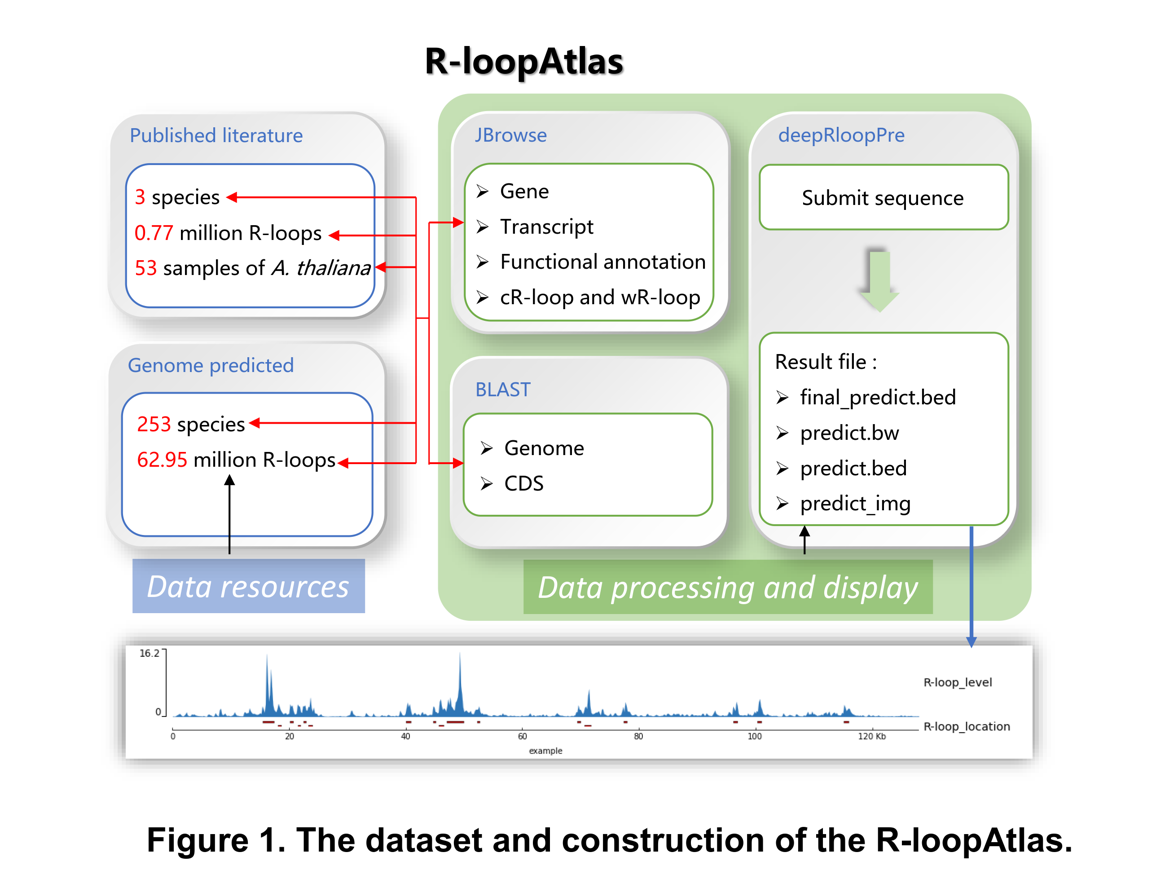
R-loopAtlas, based on a prediction tool named DeepRloopPre, is a specialized database for accessing and analyzing R-loop data. It contains the R-loop data of 254 plant species, among which the R-loop data of Arabidopsis thaliana is obtained by ssDRIP-seq, the R-loop data of Oryza sativa and Zea mays include both the data obtained by ssDRIP-seq and the prediction by deepRloopPre, and the R-loop data of other 251 species are only predicted by deepRloopPre. The main sections of our database including Home, Browse of Observed R-loop Data, Predicted R-loop Data and R-loop Atlas of Arabidopsis thaliana, Search for Observed R-loops by Gene, Observed R-loops by Region, Predicted R-loops by Gene and Predicted R-loops by Region, DeepRloopPre Online, DeepRloopPre Downloads, Predicted R-loop Data Downloads, Blast, JBrowse for Observed R-loops, JBrowse for Predicted R-loops, Publications, Tutorial, News, Links and Contact Us. R-loopAtlas v2.0 has been online on 14, April, 2022, which contains observed R-loop information of three species (Arabidopsis thaliana, Oryza sativa and Zea mays) and predicted R-loop information of 253 species.

Home
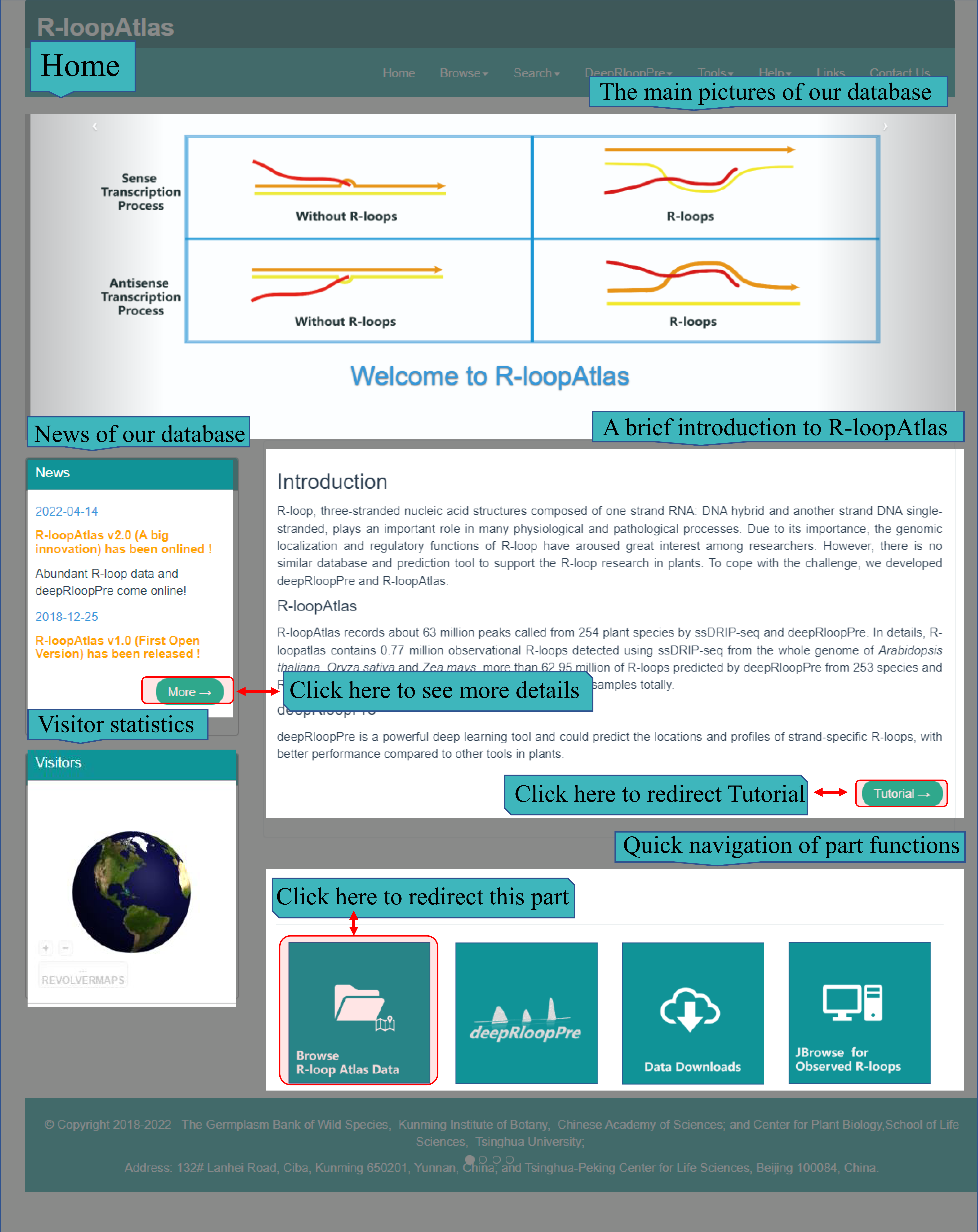
The main content of Home page is to provide a brief introduction to R-loopAtlas including the definition of R-loop, data sources and database functions, a brief history of the database, a quick access toolbar, and access statistics.
Browse
The Browse page is comprised of observed R-loop data, predicted R-loop data, and the R-loop level data in 53 samples of Arabidopsis thaliana via ssDRIP-seq and normalized by DESeq2. Its main function is to display the complete data of our database (including observed data and predicted data). And Browse provides Observed R-loop Data, Predicted R-loop Data and R-loop Atlas of Arabidopsis thaliana.
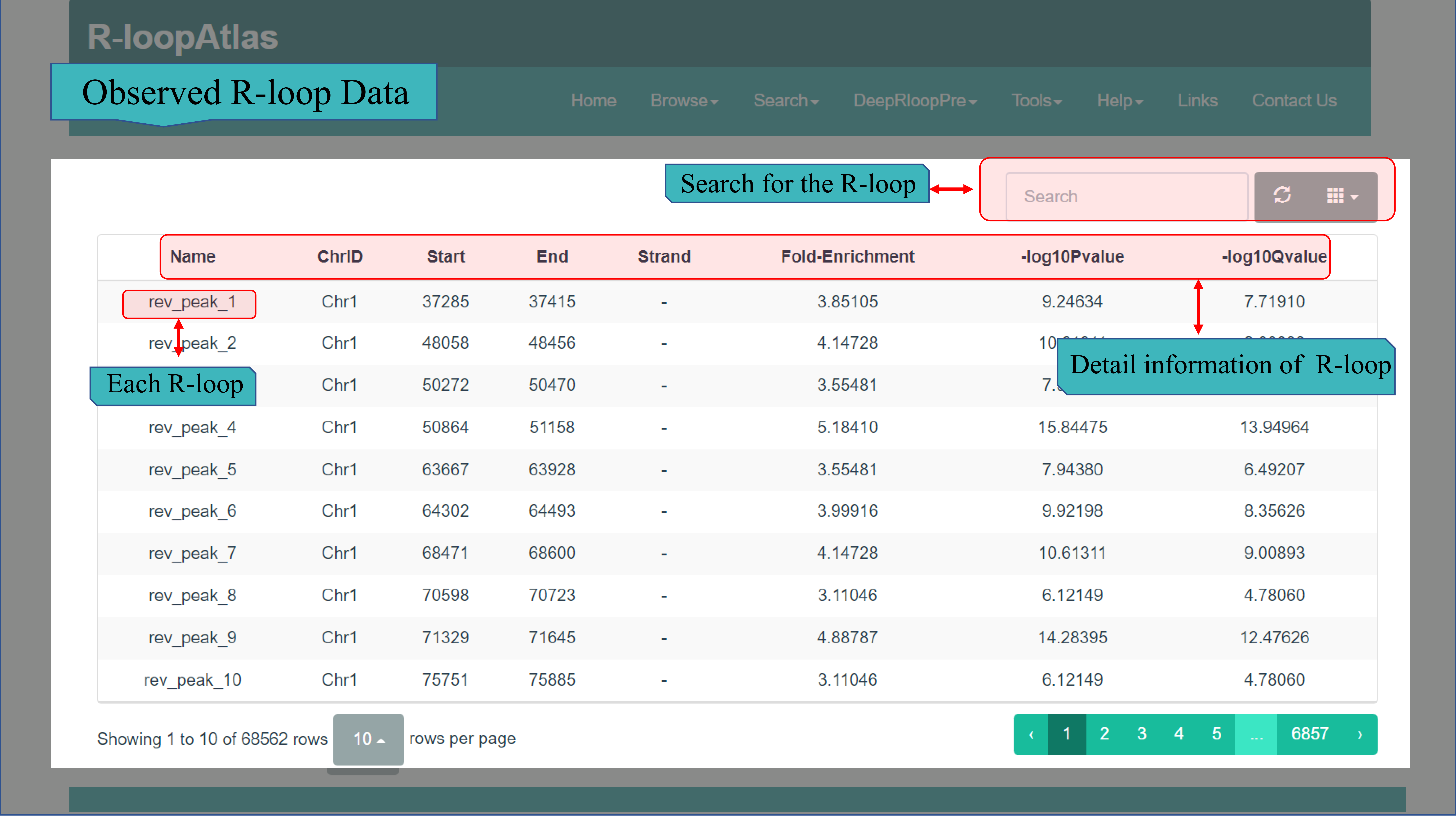
(I) The Observed R-loop Data page contains R-loop data detected from three species via the ssDRIP-seq method, including Arabidopsis thaliana, Oryza sativa and Zea mays. Statistical information for each species has also been given, both the number of cR-loop and the number of wR-loop. When click on one on specific species, detail information of each R-loop are given in the form of table, including R-loop ID, chromosome ID, R-loop start and end position, strand, fold-enrichment, P value and Q value.

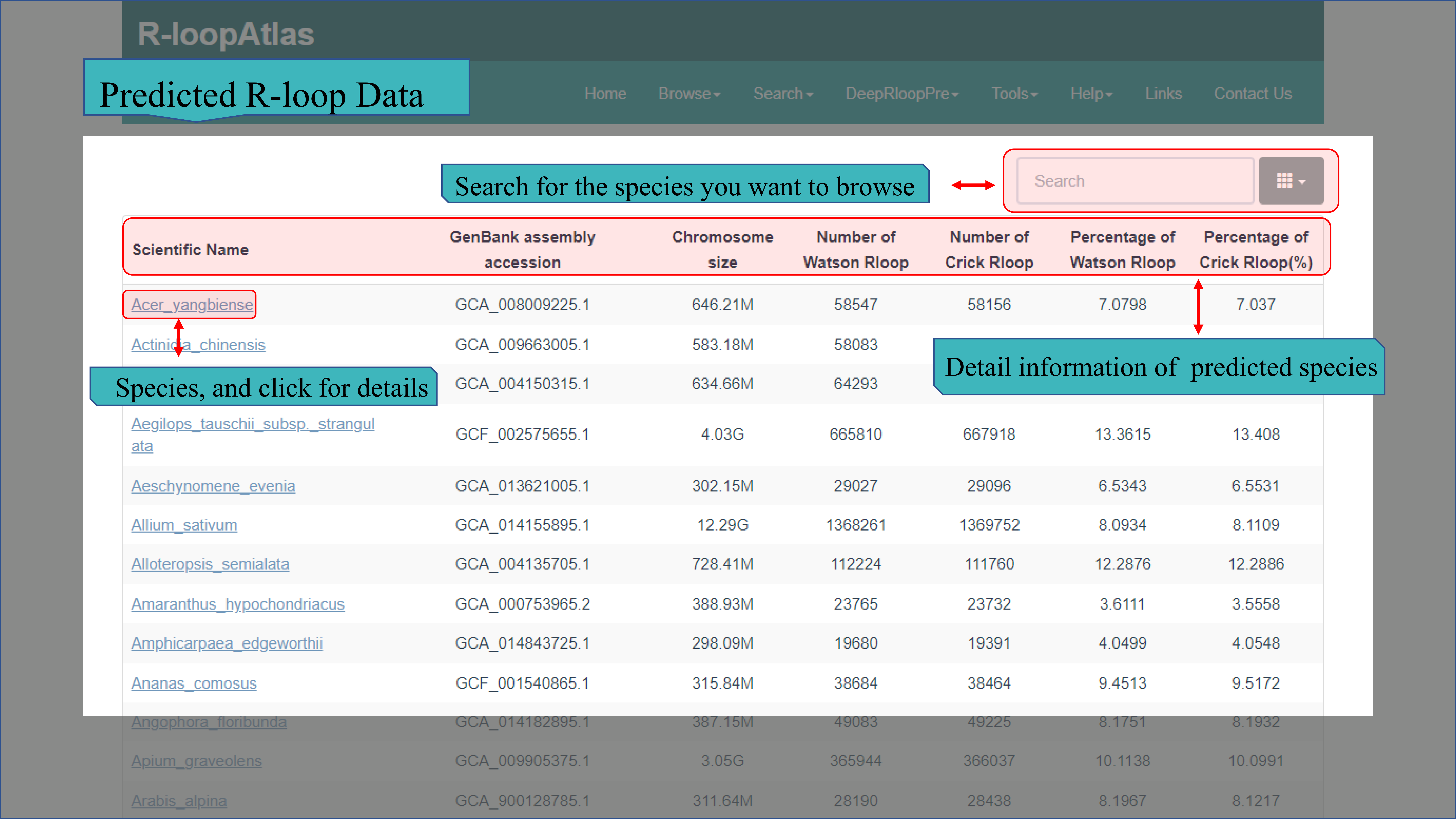
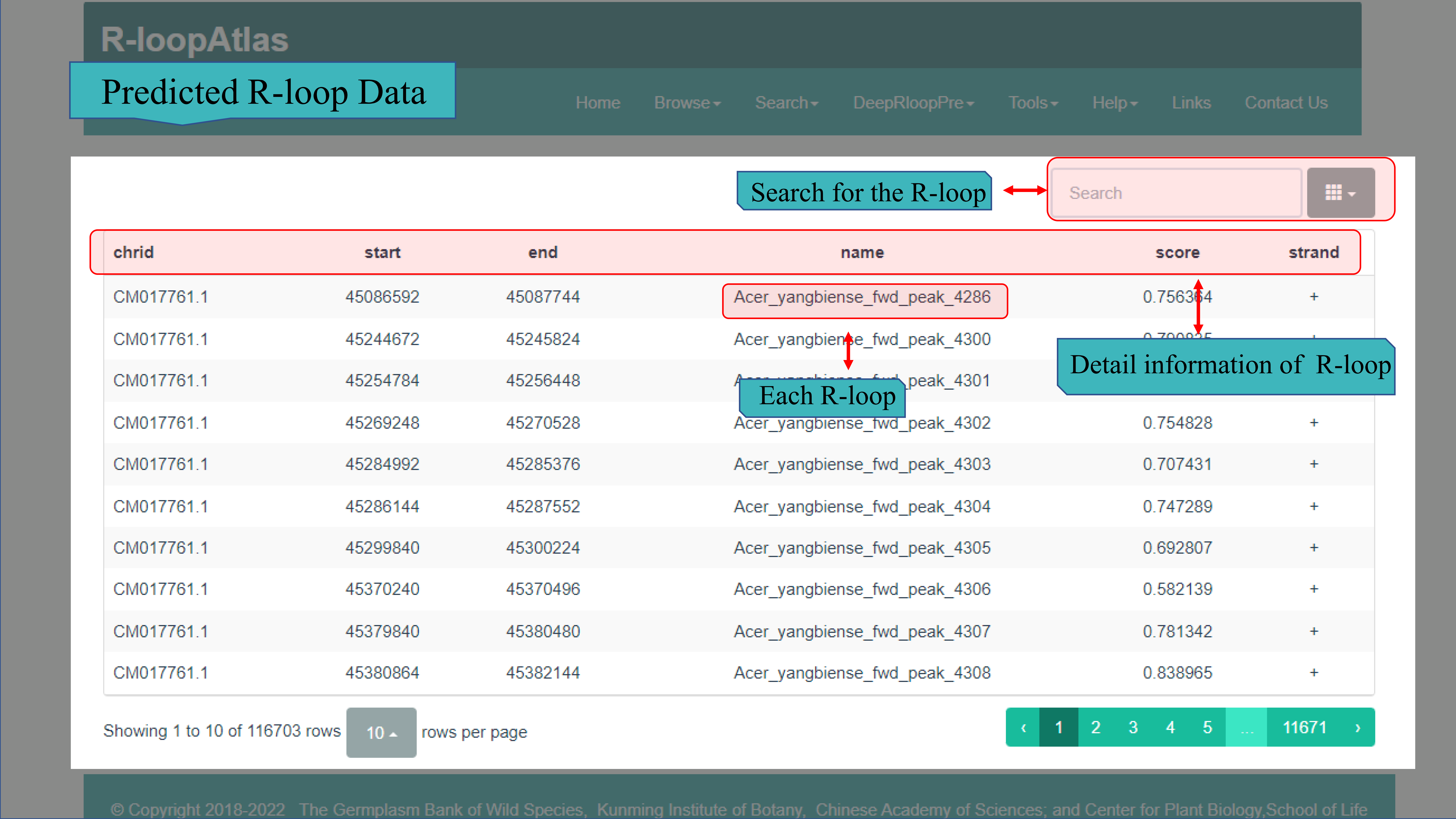
(Ⅱ) The Predicted R-loop Data page contains more than 62.95 million of R-loops predicted using deepRloopPre of 253 species except A. thaliana (A. thaliana is used as the predictive model) in R-loopAtlas. Statistical information for each species has also been given, both the Scientific Name, GenBank assembly accession, Chromosome size, number of Waston Rloop, number of Crick Rloop, Percentage of Waston Rloop and Percentage of Crick Rloop. And click on the Scientific Name, the information of every R-loop of this species is displayed, including chrid, start site, end site, name, score (means that the closer to 1, the higher the credibility) and strand.
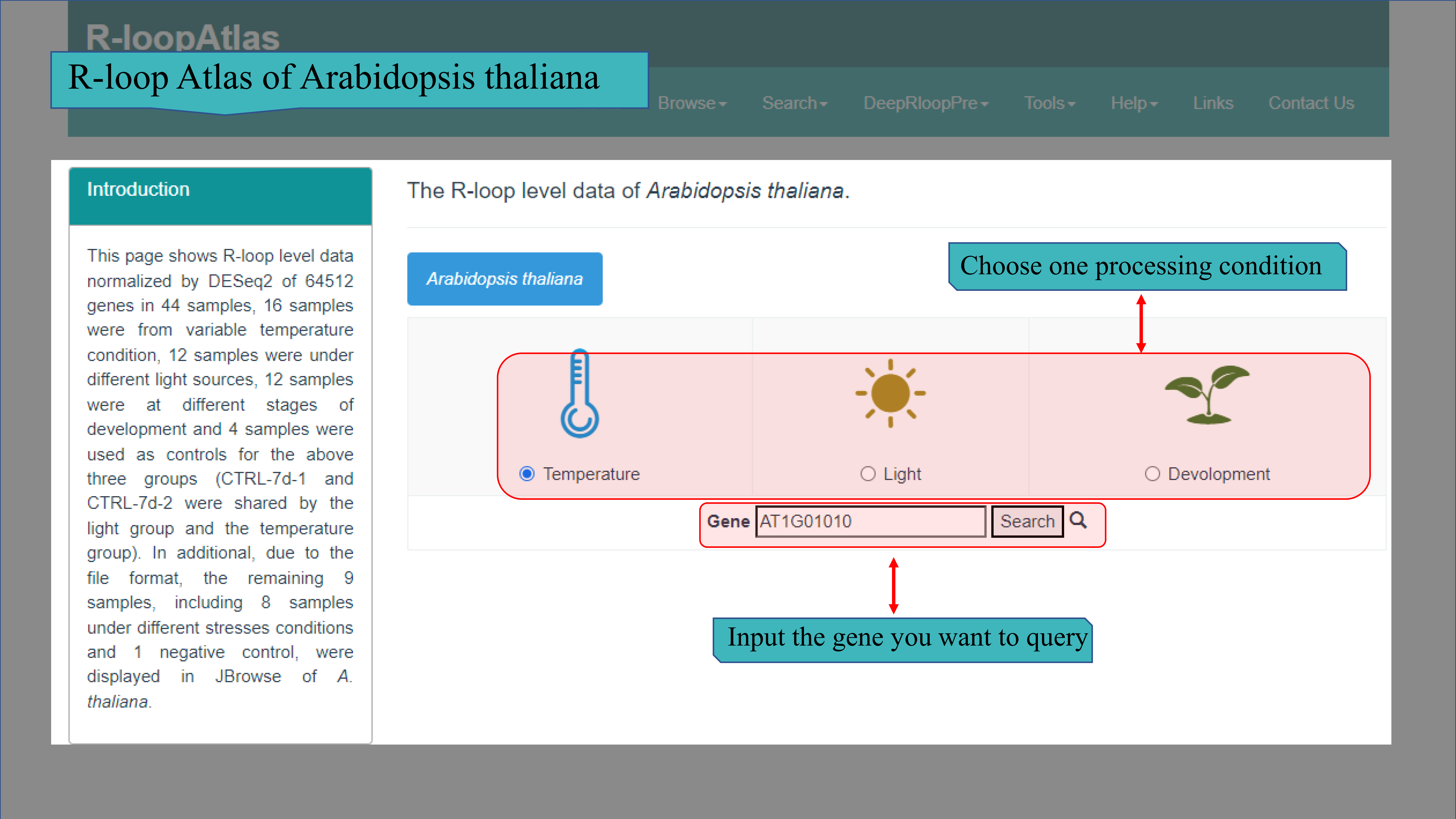
(Ⅲ) The R-loop Atlas of Arabidopsis thaliana page contains R-loop level normalized by DESeq2 of 64512 genes in 44 samples, 16 samples were from variable temperature condition, 12 samples were under different light sources, 12 samples were at different stages of development and 4 samples were used as controls for the above three experiments. These data was presented in three groups, including “Temperature”, “Photon” and “Development”. When choose one group and input the gene wanted, the relevant R-loop level will be presented in the form of bar chart and heatmap.At the same time, it will list the condition involves the related samples.

Search
The function of Search page is to receive users' specific search needs and provide with corresponding results. Based on the data type and search requirements, Search provides search Observed R-loops by Gene, Observed R-loops by Region, Predicted R-loops by Gene and Predicted R-loops by Region. And due to some speceis are not well-annotated, only 121 species can be searched in this part.
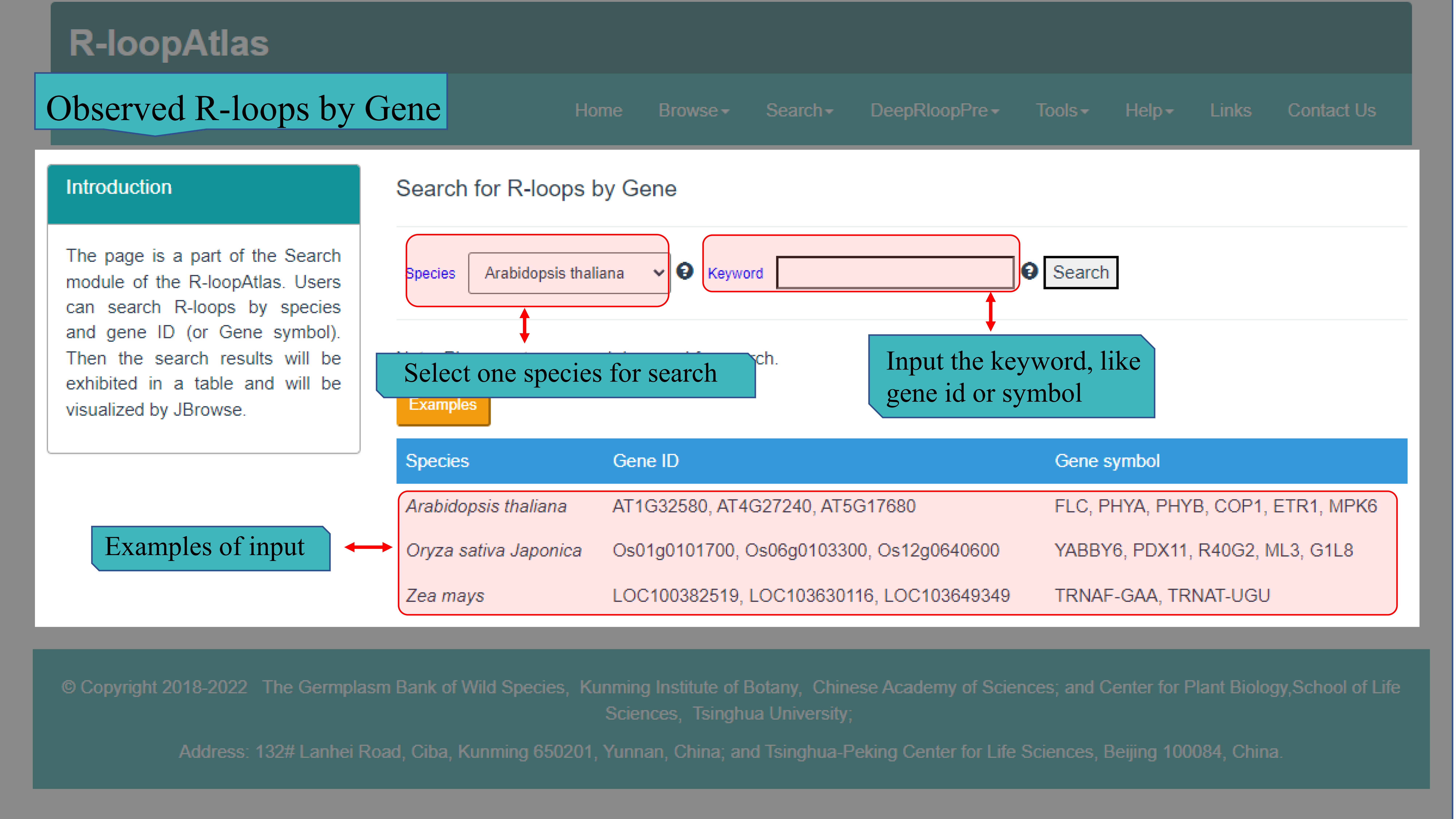
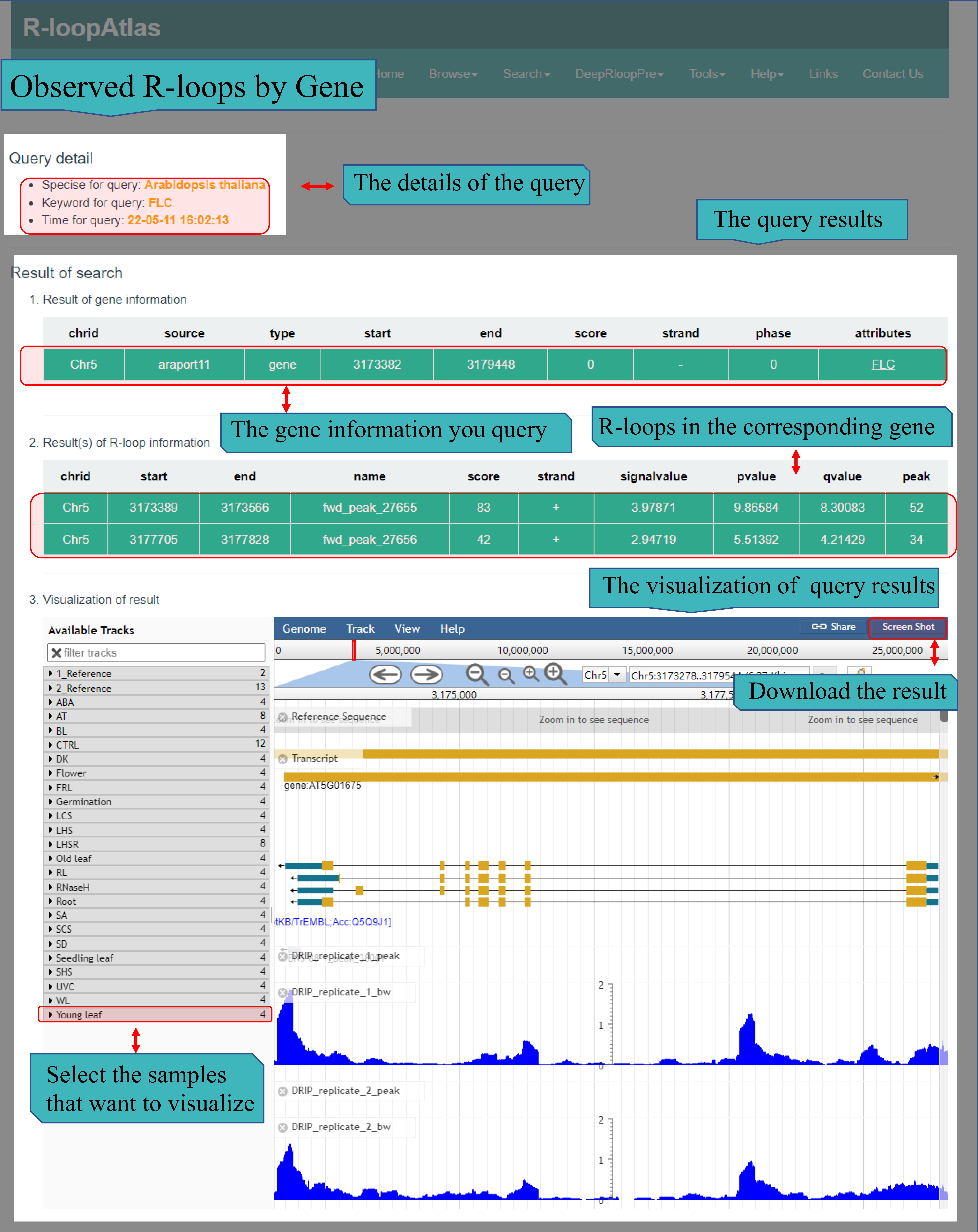
(I) The Observed R-loops by Gene page main provides search function for three species (Arabidopsis thaliana, Oryza sativa and Zea mays ) that used to generate R-loops via ssDRIP-seq. Users can select the desired species from the selection box and enter keywords such as Gene ID and Gene symbol in the "Keyword" box. After the server received the submitted search, the results will be returned to the web page in HTML, including query details, detailed information of target gene, R-loop details corresponding to target gene (or region), and visual display of target gene and R-loops in JBrowse.
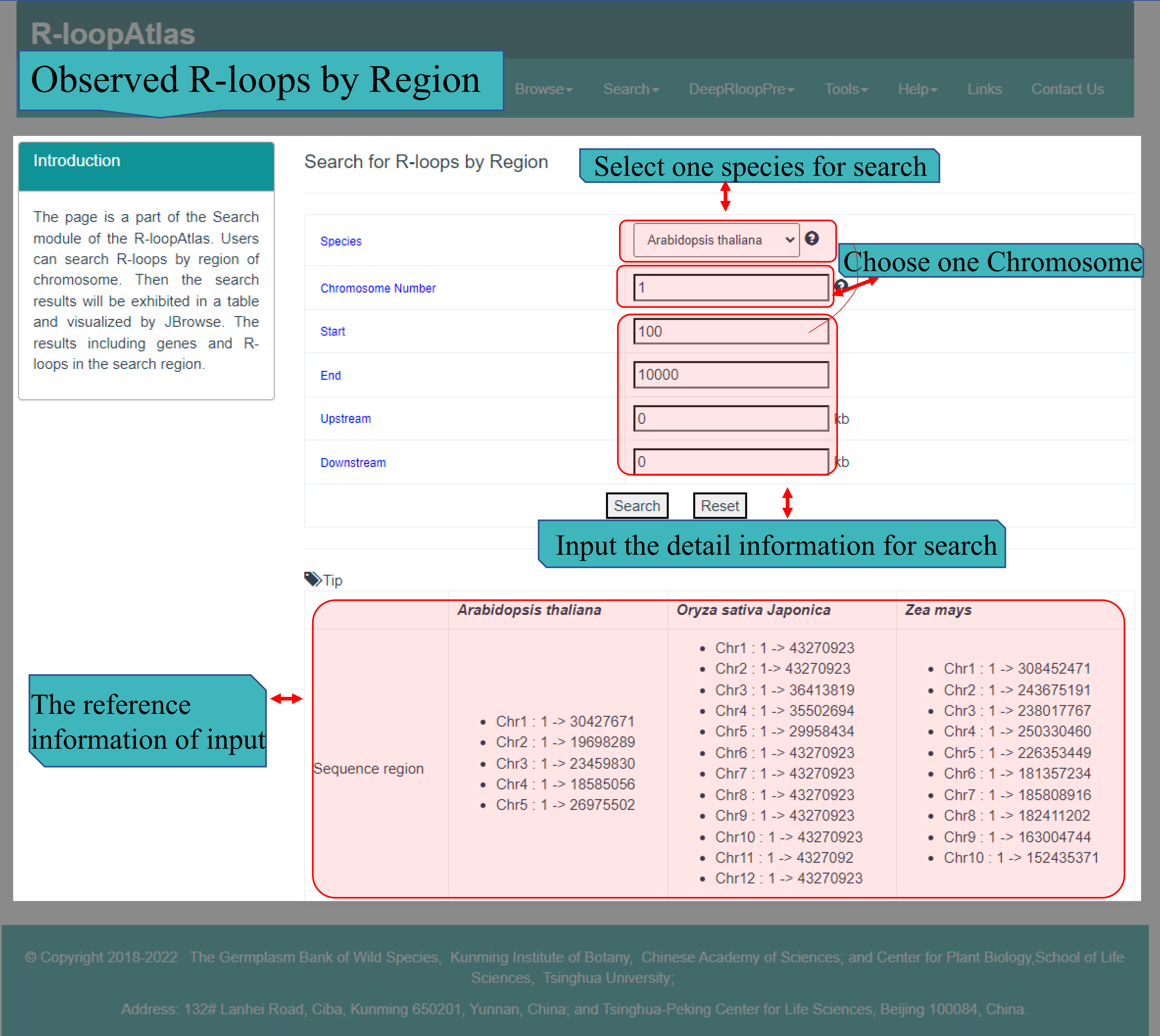
(Ⅱ) The Observed R-loops by Region page main provides search function for three species (Arabidopsis thaliana, Oryza sativa and Zea mays ) that used to generate R-loops via ssDRIP-seq. Users can select the species from the drop-down list, input the chromosome number, enter the start and end location, and input extended display upstream and downstream of the target area (or keep the default zero, which means no extended display). After the server received the submitted search, the results will be returned to the web page in HTML, including query details, detailed information of target region, R-loop details corresponding to target region, and visual display of target region and R-loops in JBrowse.
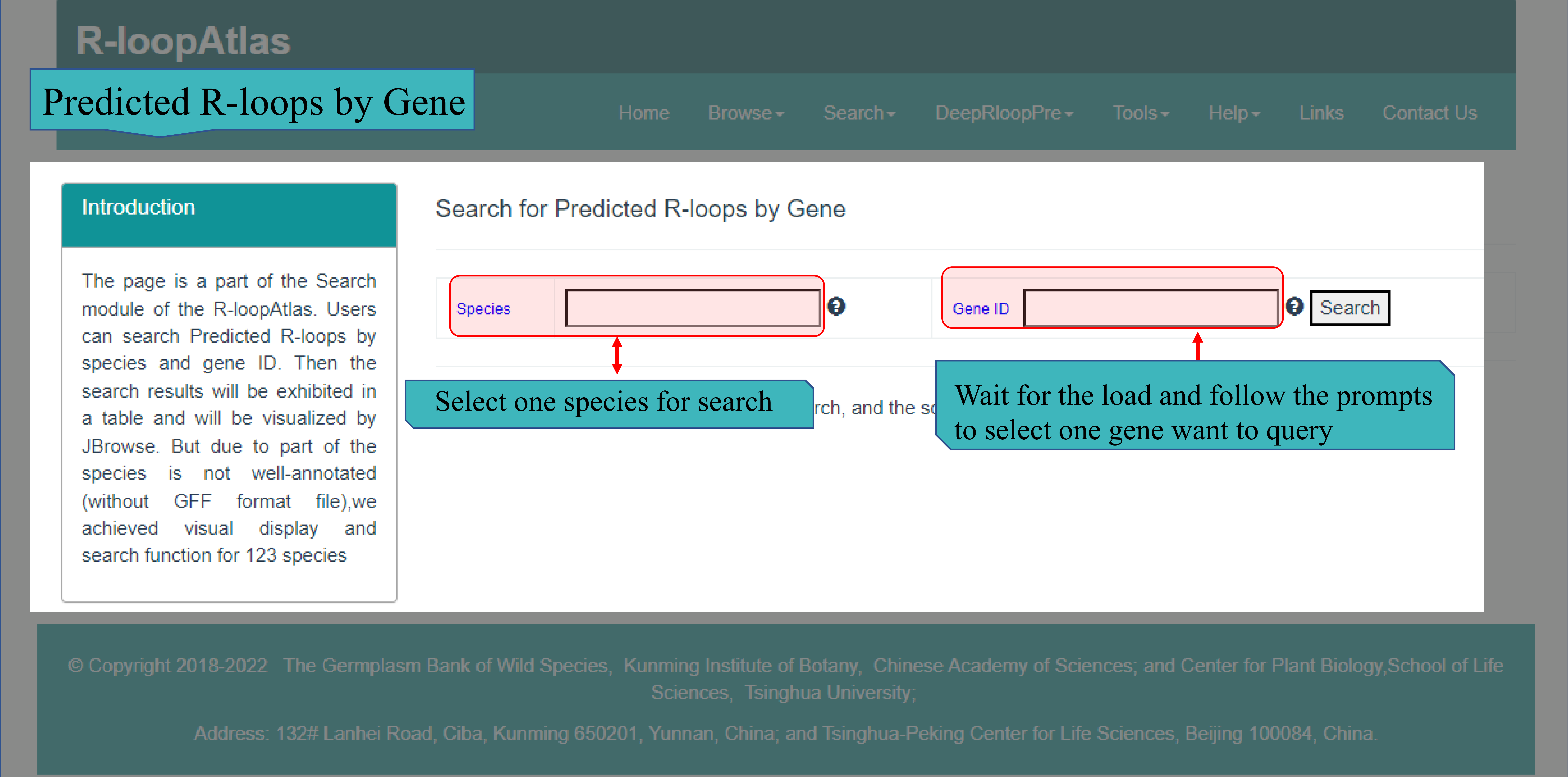
(Ⅲ) The Predicted R-loops by Gene page main provides search function for 120 species that used to generate R-loops by using deepRloopPre. Users can select the desired species from the selection box and enter the Gene ID. After submitted, the results will be returned to the web page in HTML, including query details, detailed information of target gene, R-loop details corresponding to target gene (or region), and visual display of target gene and R-loops in JBrowse.
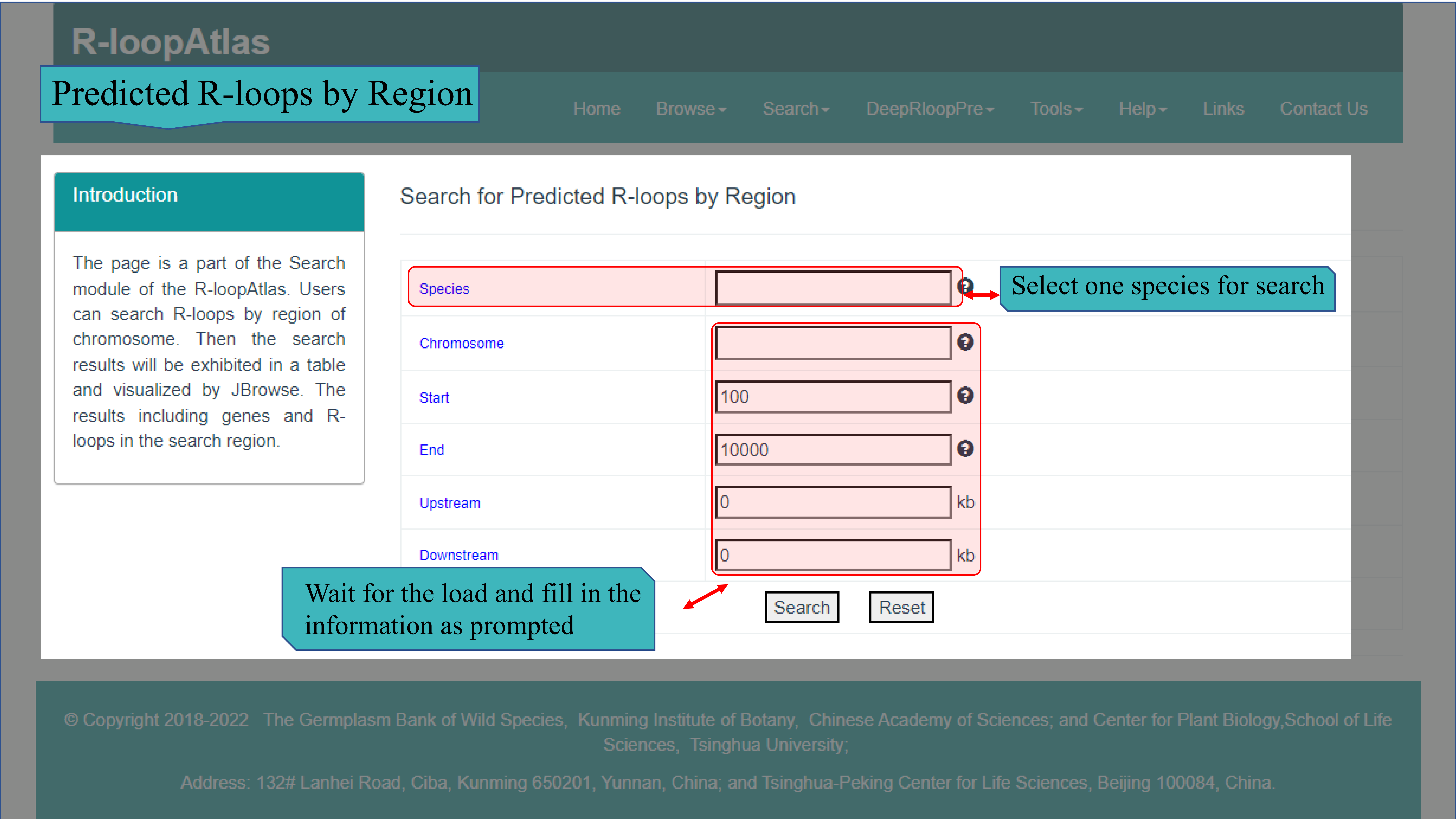
(Ⅳ) The Predicted R-loops by Region page main provides search function 120 species that used to generate R-loops by using deepRloopPre. Users can select the species from the drop-down list, input the chromosome ID, enter the start and end location, and input extended display upstream and downstream of the target area (or keep the default zero, which means no extended display). After the server received the submitted search, the results will be returned to the web page in HTML, including query details, detailed information of target region, R-loop details corresponding to target region, and visual display of target region and R-loops in JBrowse.
DeepRloopPre
DeepRloopPre, a powerful R-loop predicted tool based on deep learning, could predict the locations and profiles of strand-specific R-loops, with much better performance compared to other tools in plants. And deepRloopPre is also uesd to generate large amounts of predictive R-loop data we showed is also integrated into our database. We also provide DeepRloopPre Online, DeepRloopPre Downloads and Predicted R-loop Data Downloads.
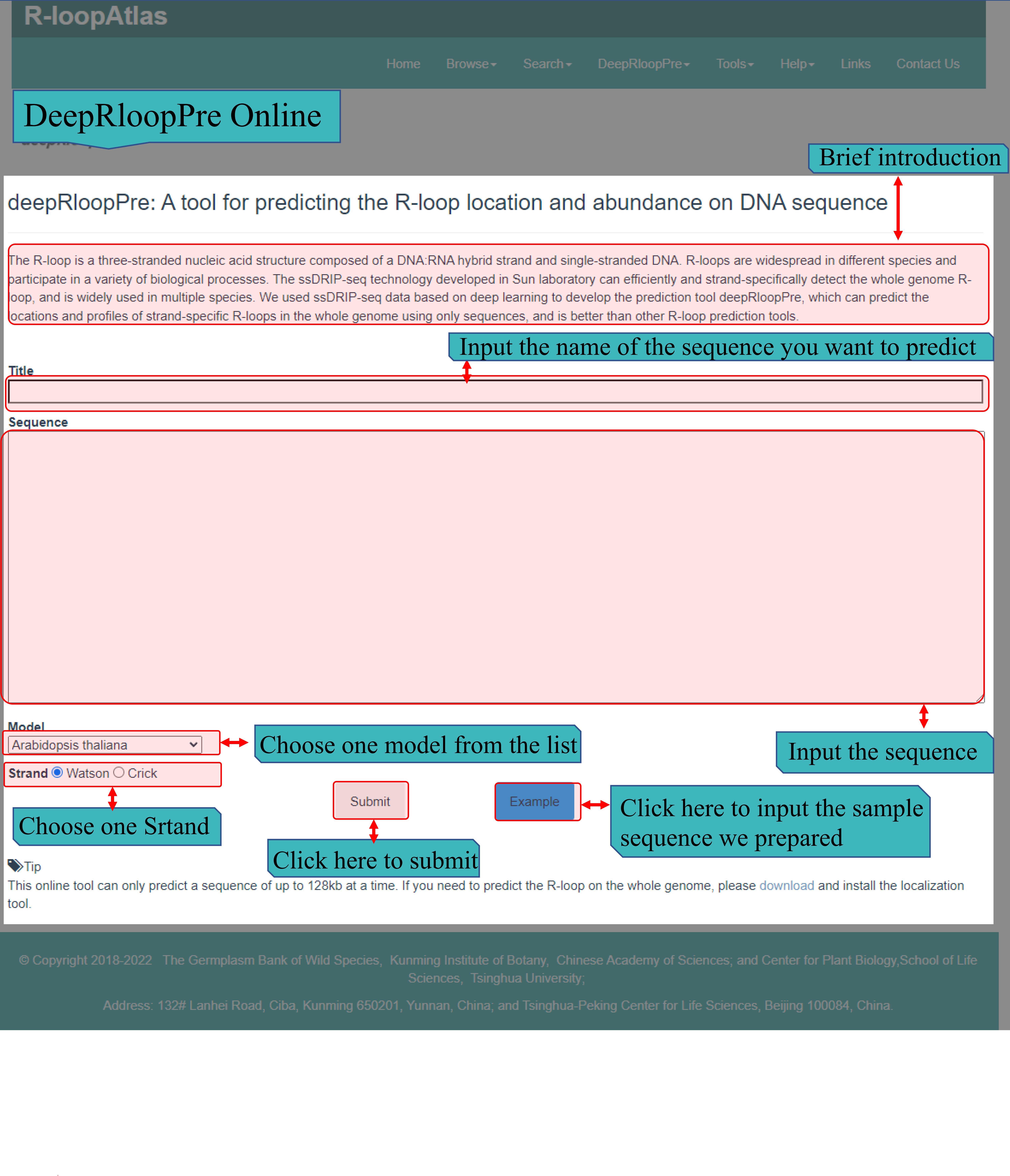
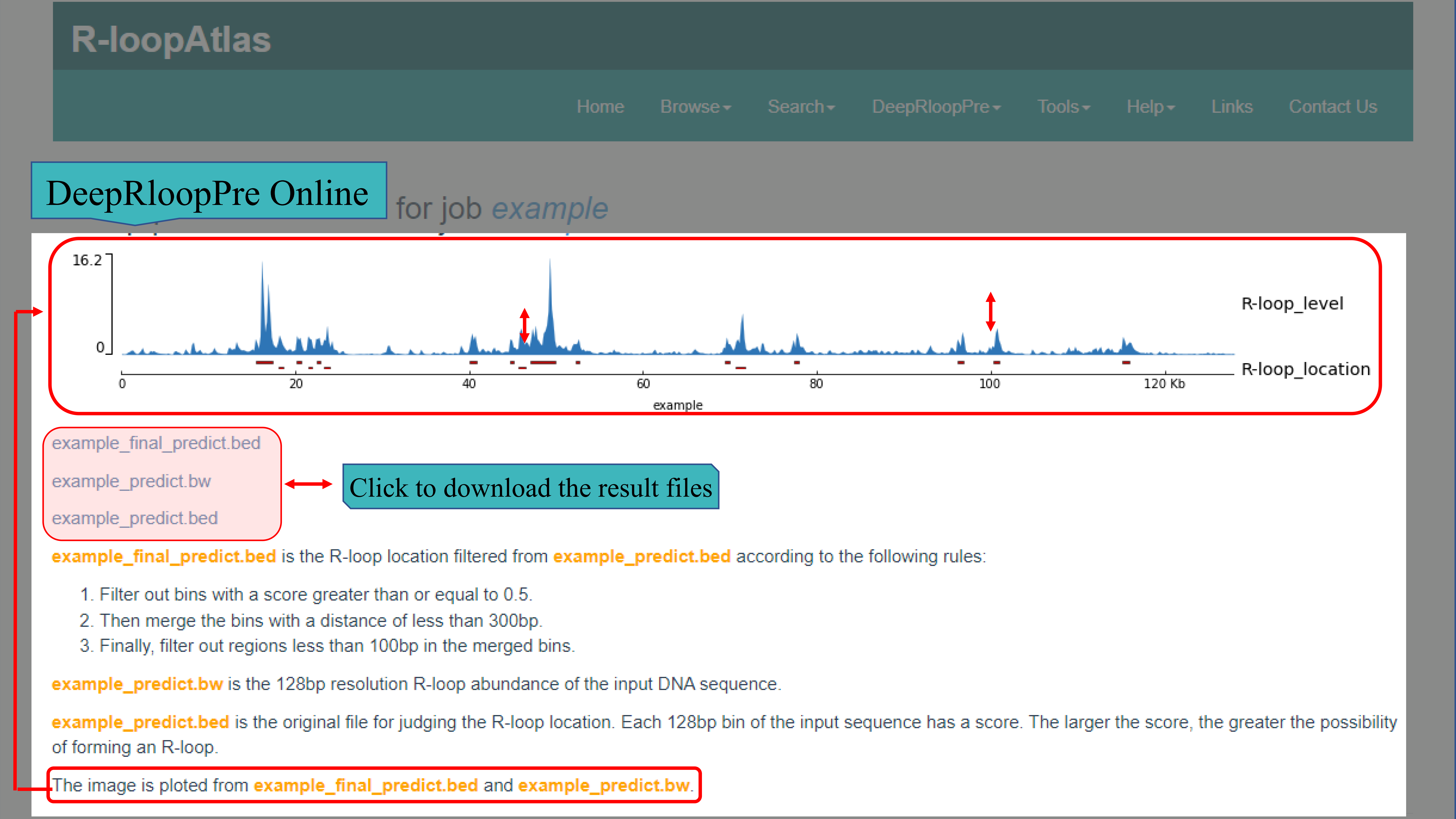
(I) DeepRloopPre Online is a user-friendly interface that can run directly on the website. When using it, user only needs to input the title and sequence, select one appropriate model and strand. After submitting, the results including one BW file, two BED files and an PNG image according to _final_predict.bed and _predict.bw file which plotted by the submitted sequences. Please note that the sequence predicted must be no more than 128kb and no less than 1kb. And if need to predict the R-loop on the whole genome, you can download and install the localization tool which we also provided.

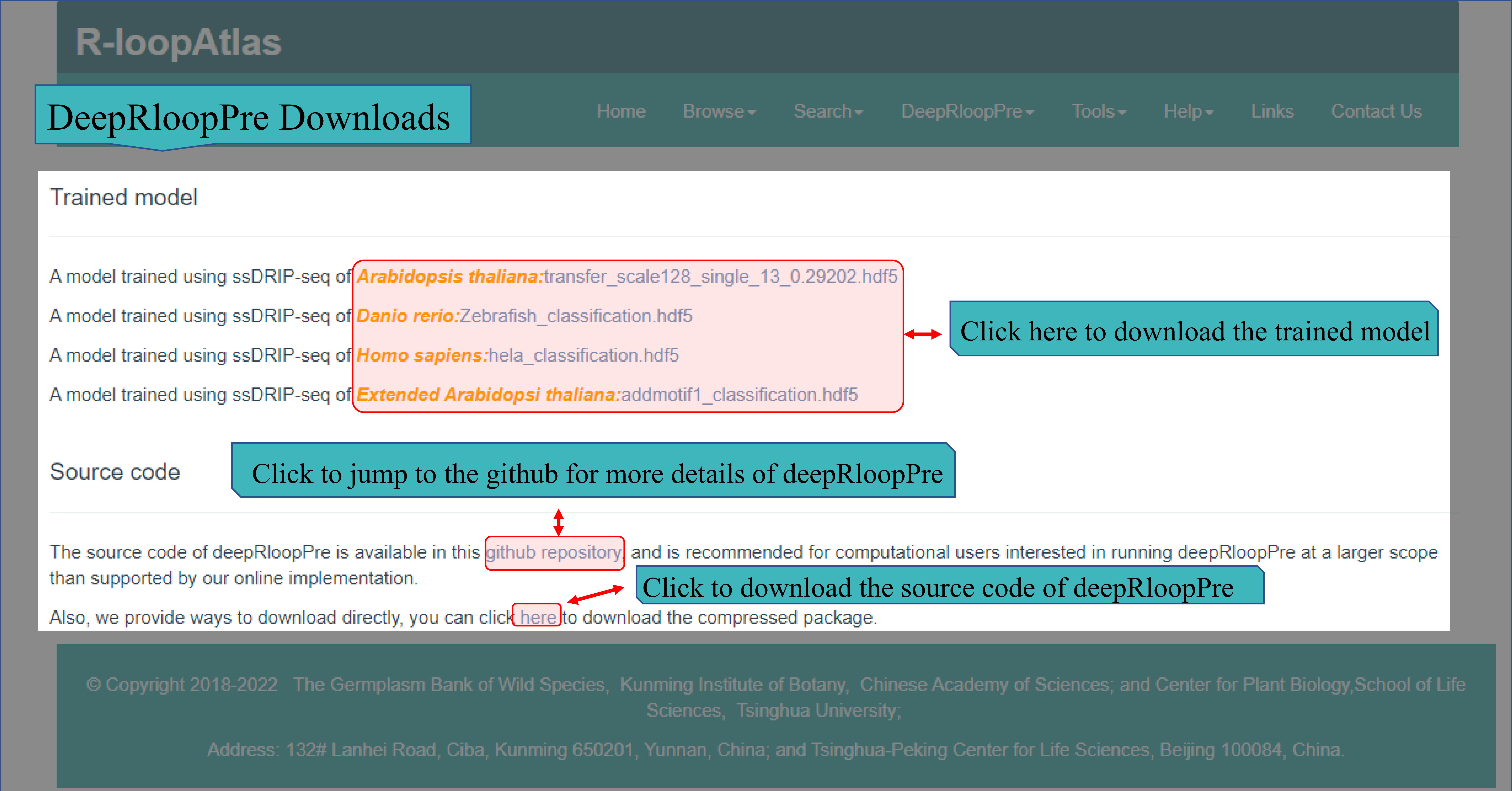
(Ⅱ) The DeepRloopPre Downloads pages provides source code and trained model of deepRloopPre. User can run deepRloopPre at a larger scope when needed. And in this page, we also provided link of github repository.
(Ⅲ) The Predicted R-loop Data Downloads page provides open-access link of the predicted R-loop data. when downloading the data, please click on the scientific name of the species.

Tools
The Tools page provides bioinformatics tools that maybe used in our R-loopAtlas, like blast and JBrowse. This part mainly includes Blast, JBrowse for Observed R-loops and JBrowse for Predicted R-loops.
(I) The BLAST page is formed by Viroblast which is a standalone Blast web tool that integrated into our database, is commonly used in the sequence similarity search. And its database contains sequences of geneome and cds of all species involved in our R-loopAtlas.

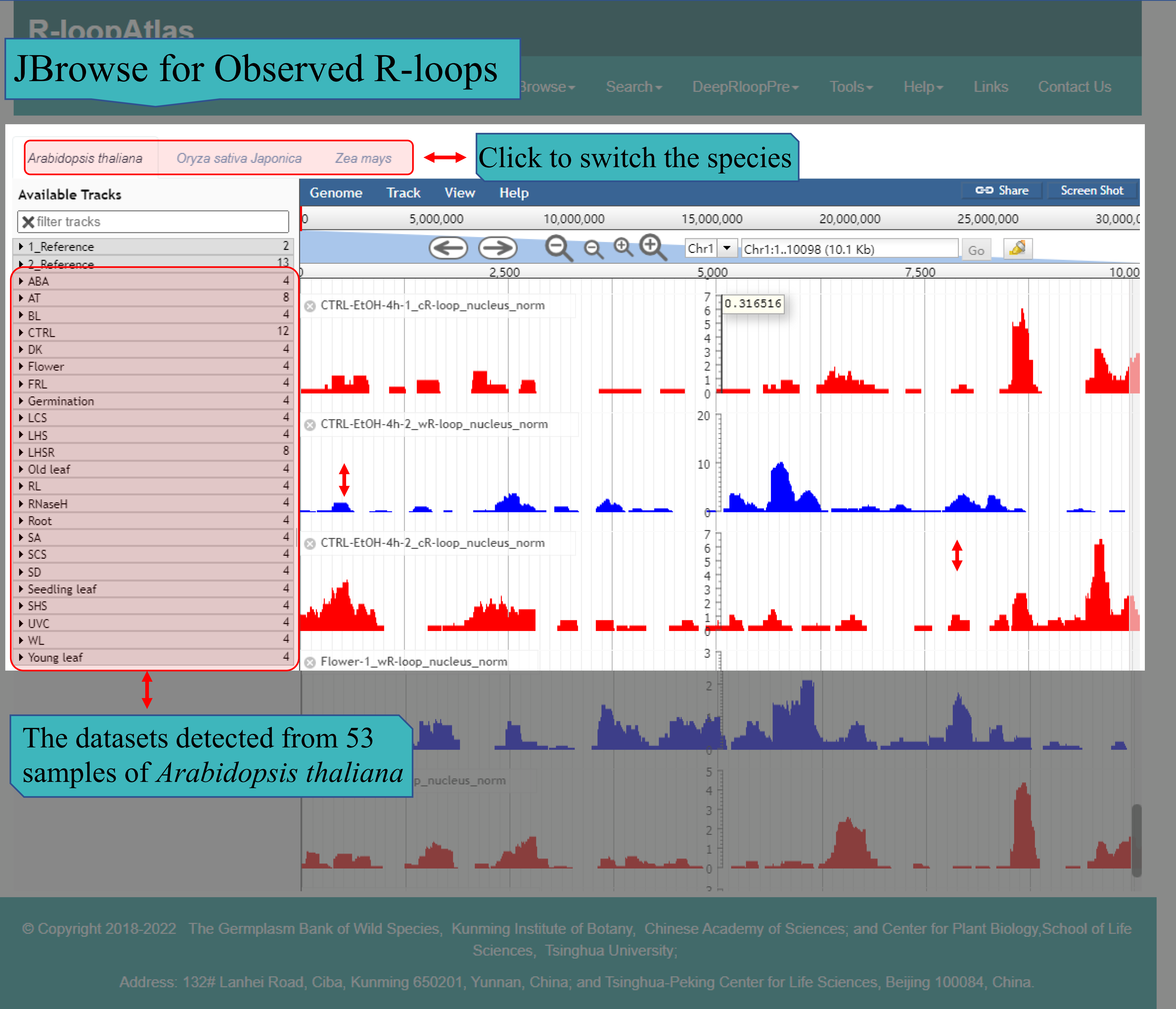
(Ⅱ) The main function of JBrowse for Observed R-loops page is to provide the visualization of R-loops observed via ssDRIP-seq from the three species independently, including Arabidopsis thaliana, Oryza sativa and Zea mays. For Arabidopsis thaliana, R-loop level data of all 53 samples are presented at this page.
(Ⅲ) The main function of JBrowse for Predicted R-loops page is to provide the visualization of R-loops predicted by deepRloopPre from other 253 species. For convenience, the 253 species are divided into 20 groups according to the phylogenetic relationship. And JBrowse contains the basic sequence and BW format files of R-loop data for each species.

Help
The Help page provides some public information of our R-loopAtlas. This part mainly includes Publications, Tutorial and News.
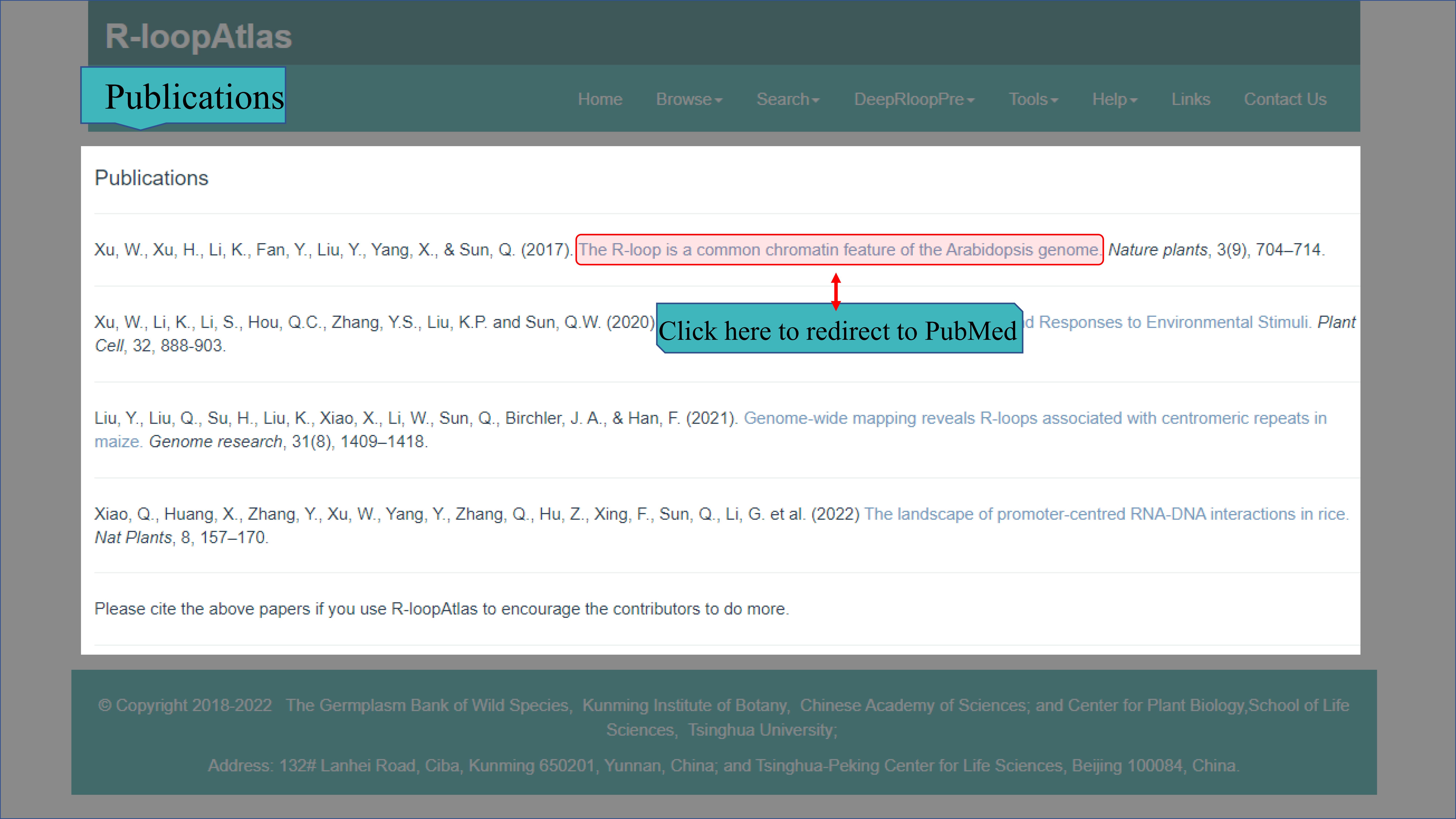
(I) The Publications page mainly lists the relevant literature which provide part of the data in our R-loopAtlas.
(Ⅱ) The News page records the development history and important release versions of our database.
Links
The Links page provides some website addresss of related databases about R-loops and bioinformatics tools uesd in our database.
Contact Us
The Contact us page introduces the leader of the project and the main system development personnel. If you have any questions about our database, you can send back to us via email. We will reply in a timely manner.
